Studying interactions between myeloid cells and their microenvironment
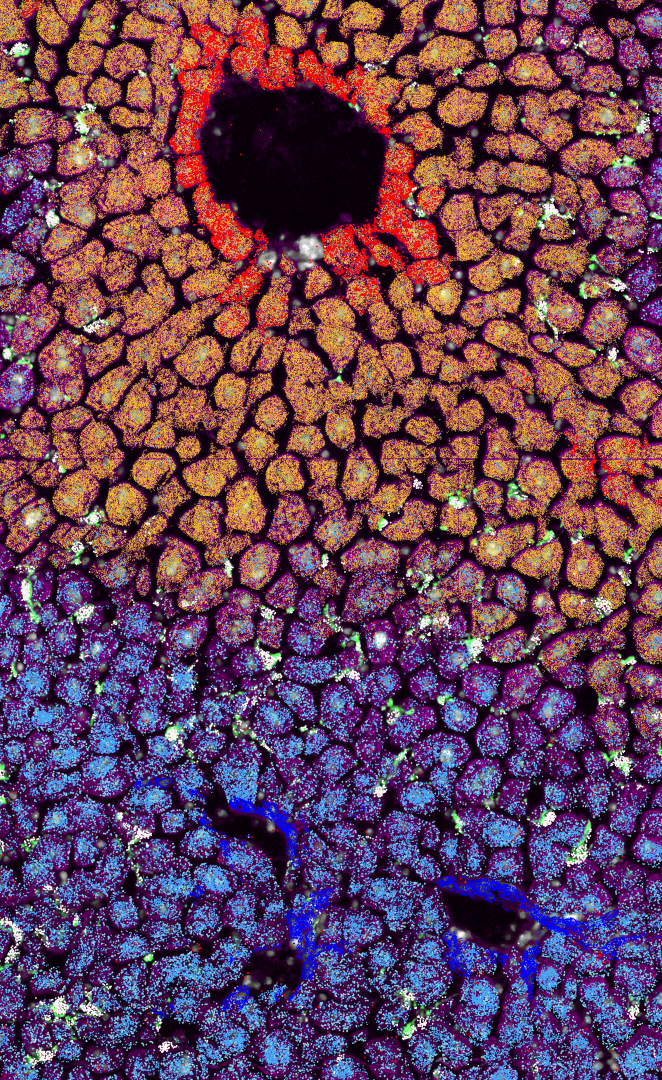
We study the interactions between macrophages and their microenvironmental niche. We invest heavily in these technologies: a) spatial multi-omics, b) AI-driven spatial analysis, c) epigenetics, d) in vivo CRISPR screens. Each project involves human and murine liver samples. The Guilliams lab is member of the Immgen (www.immgen.org) and Human Cell Atlas (www.humancellatlas.org) consortia. Our liver cell atlas work revealed the modular architecture of the liver sinusoids, which are constituted of hepatocytes, sinusoidal endothelial cells (LSECs), stellate cells and Kupffer cells (KCs). We found that each KC projects an important part of its body across the LSEC barrier to be in close contact with a stellate cell and a hepatocyte, generating a four-cell-circuit. The genetic program of these four cells is completely integrated, with each cell representing an essential part of the cellular niche of the other cells. This suggests that throughout evolution hepatocytes, LSECs, stellate cells and KCs have become genetically integrated to function together as an interconnected functional sinusoidal module. This modular view forms the basis of our research where we aim to (i) unravel the cell-cell interactions forming the blueprint of the homeostatic sinusoidal liver module, (ii) unravel the signals driving multiplication of liver modules during neonatal liver growth and upon regeneration, (iii) understand how metastatic cells hijack the liver building blocks to build their own modules.
Areas of Expertise
- Macrophages
- Innate immune system
- Tissue Regeneration
- Liver Metastasis
- Single-cell and Spatial multi-omics
Technology Transfer Potential
- Hepatology
- Myeloid cells
- Single-cell and Spatial multi-omics
- in vivo CRISPR screens
- AI-driven spatial analysis
Selected publications
- Guilliams, M. et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 185, 379-396 (2022). Visit ➚
- Ydens, E. et al. Profiling peripheral nerve macrophages reveals two macrophage subsets with distinct localization, transcriptome and response to injury. Nat Neurosci 23, 676-689 (2020). Visit ➚
- Bonnardel, J. et al. Stellate Cells, Hepatocytes, and Endothelial Cells Imprint the Kupffer Cell Identity on Monocytes Colonizing the Liver Macrophage Niche. Immunity 51, 638-654 (2019). Visit ➚
- Scott, C. L. et al. The Transcription Factor ZEB2 Is Required to Maintain the Tissue-Specific Identities of Macrophages. Immunity 49, 312-325 (2018). Visit ➚
- van de Laar, L. et al. Yolk Sac Macrophages, Fetal Liver, and Adult Monocytes Can Colonize an Empty Niche and Develop into Functional Tissue-Resident Macrophages. Immunity 44, 755-768 (2016). Visit ➚
Bibliography
- Full bibliography Visit ➚